Challenges in the Application and Design of NGS Capture Probes
In NGS whole-genome sequencing, the massive data presents significant challenges in analysis. however, if you are interested of certain region on genome or disease ,targeted capture of the regions and sequencing is a better choice .This method offers lower sequencing costs, higher sequencing depth, and higher data efficiency, making it suitable for detecting mutation types such as SNP, InDel , CNV and Fusion.
There are mainly two methods for targeted capture:
1. Amplicon Sequencing based on multiplex PCR:
Multiplex PCR is an effective method that uses primer-based amplification to obtain gene fragments. however, the number of amplicons in a single experiment is limited and the design is difficult
2. Target Enrichment based on probe:
Biotinylated probes capture target gene fragments through hybridization to target-specific biotinylated probes ,which are then isolated by magnetic pulldown. The target fragments are sequenced .
Compared to multiplex PCR, Hybridization-based enrichment is a useful strategy for detect wider variety of variants type, target a higher amount of total gene content, a higher tolerance for mismatches in the probes.
Among these methods, the design of capture probes with good specificity and uniformity is crucial for hybridization capture. Many factors need to be considered, including GC content, probe structure, and location. Therefore, a good probe design software is essential!
BiOligo capture probe design
In order to address the challenges of capture specificity and uniformity in hybridization capture probe design, Bioligo NGS team has developed the advanced hybridization capture probe design tool, Smartbaits,with results of the experiments, which is able to complete the probe design easily, efficiently and with high quality.
Advantages:
1. Multiple Recognition Methods: Smartbaits can recognize various common input formats: bed format files, Fasta files, Gene Symbols, etc.
2. Optimized Algorithms: The algorithms in Smartbaits consider probe GC balance, specificity, homologous and heterologous dimerization between probes, hairpin structures, etc. It employs a finer granularity to find the optimal probe solution while ensuring the maximum coverage with fewer probes, thereby reducing costs.
3. More Flexible Parameter: Users can choose different density modes such as 1x, 2x tiling, and overlap. They can also select different reference genomes or target sequences, not limited to model organisms or the human genome. Smartbaits also support probe risks based on sequence specificity, extreme GC, and secondary structure regions.
4. Flexible Design Schemes: Different intervals can employ different design schemes. For example, gene intervals and intergenic regions, SNP frameworks, and MSI utilize different optimization algorithms for design.
5. Sustainability: Through spike-in methods, additional probes can be added to existing panels to form new panels.
6. Flexible and Low Threshold Usage: Smartbaits uses SQL relational database back-end to minimize issues with high memory load and extensive I/O computation time for data files. This allows users to quickly reanalyze with multiple different filtering criteria and minimizes the usage threshold.
Design Principle:
Different regions are designed using different methods.
1. Design for Common Regions: Based on the principle of central symmetry, the algorithm searches for the optimal probe set within a movable window range.
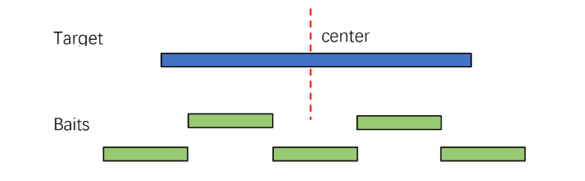
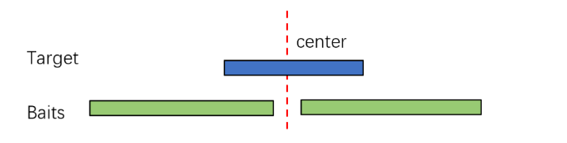
2. MSI Region Design: Most microsatellite sequences are short with high repeat content. Conventional designs often fails to achieve desired capture results. Therefore, an optimized strategy is employed using paired-end probes to cover both ends of the target region separately.
Design Process
1. Input:
Accept user input in .bed file format, genome.fasta, or gene symbol. The default design is 1x tiling, but users can adjust it to 2x tiling if needed. The program will analyze the input information and save it in the database.
2. Input Quality Check:
Convert input file into FASTA sequences for quality check analysis. including analyzing sequence GC content, Tm, sequence complexity, etc. The analysis results will be saved to the SQL database.
3. Probe Design:
Apply sliding window segmentation to candidate target regions. Generate all possible probes with a length of 120 bp(probe length), following the principle of symmetry from left to right with a step of 1.
4. Probe Screening:
Analyze all probe sets and perform quality checks. Each probe performed specificity analysis using blastn. decision tree is used to filter probe sets based on user-defined parameters, focusing on sequence specificity, GC content, Tm, secondary structure, etc., to obtain the final probe set.
5. Report Design:
After design completion, probes are classified into risk levels: "green" represents safe probes ensuring capture efficiency and specificity, "yellow" indicates some specificity risk, and "black" signifies poor specificity or capture efficiency, not recommended for addition.
Here are the final three sets of results:
1. Probe Design Report: This PDF report contains information such as probe coverage and the number of probes designed.
2. Pass Baits Report: This report includes information on all qualified probes, along with the quality check data for each probe.
3. All Baits Report: This report contains information on all probes, including black and yellow probes, along with their quality check data.
Performance Comparison
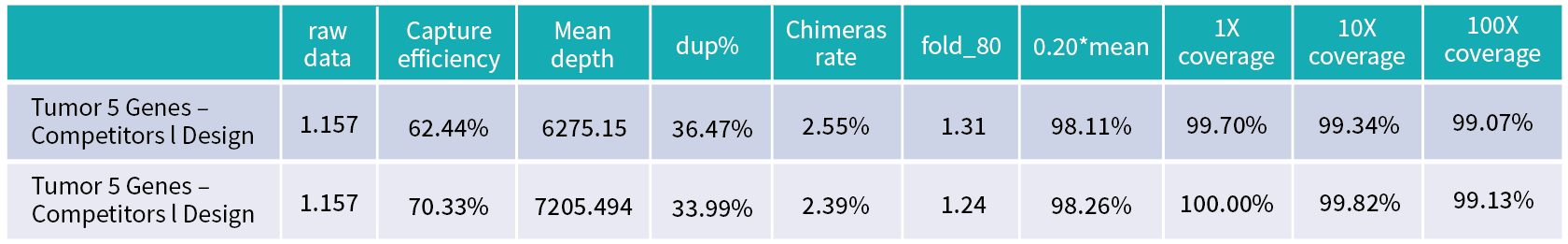
Below is a comparison of performance between using Smartbaits from BiOligo and Ixx Company for designing probes targeting genes KRAS, ALK, ROS1, EGFR, and BRAF (CDS region), followed by hybridization capture experiments. When downsampling to same amount of data, BiOligo achieved higher capture efficiency, higher average sequencing depth, lower fold80, and reached 100% 1x coverage.
The application scope
The NGS Hybridization-based enrichment can be applied to various types of research , it allows researchers the ability to reliably sequence exomes or large numbers of genes ,such as Human whole-exome sequencing, cancer and genetic disease research, complex disease diagnosis, Custom genomic sequencing,etc.
The usage method
You can reach out to us for design through email at blg-mkt@bioligo.com, private messaging on our public account, or contacting our local sales representatives. Smartbaits will also be available on the Bioligo official website in the future, so please stay tuned!




